Bacterial dormancy
Microbial dormancy as a model of a natural response to changing environmental conditions
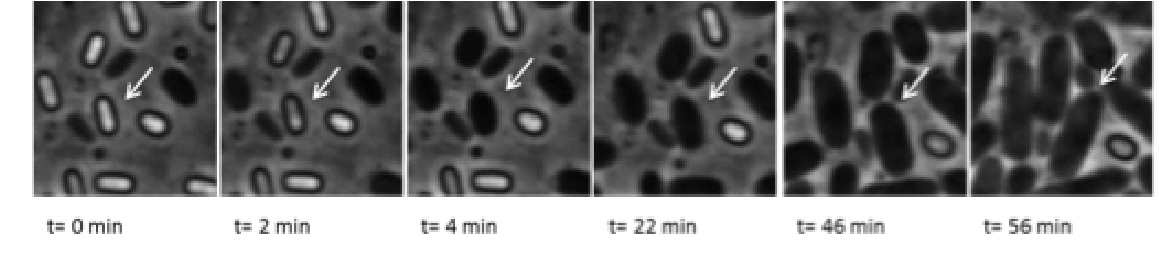
To cope with various stresses, microorganisms have evolved various strategies to withstand environmental conditions that limit active growth and plunges them in a viable but non-growing or dormant state to endure the harsh stress and resume growth once the stress subsides. Dormancy often involves the formation of a specialized morphotype, referred to here as a dormant cell (DC) (also known spore or spore-like cell).
Studying DCs in selected genetically tractable models has provided the foundation for understanding the genetic bases of DC development, and subsequently allowed the biochemical characterization of the underlying mechanism. However, DC studies are not only important for understanding mechanisms of differentiation and their regulation in response to environmental cues, but it also as recently shown to provide insight into when and how the breakpoint between monoderm (classically called Gram-positive) bacteria and diderm (Gram-negative) bacteria occurred in evolution. Prior studies claimed that DCs are restricted to a few bacterial clades. However, given the diversity of microbial ecosystems, there is a significant chance that alternative pathways based on different genes and generating uncharacterized DCs remain to be discovered. Indeed, we recently found evidence for spore-like DCs in five strains belonging to additional bacterial clades. In two of these clades, the production of the spore-like (phase bright) DCs is based on novel differentiation pathways and underlying genes. The five strains were all collected at geothermal locations, which are habitats highly selective for the ability to form DCs.
We will use these new strains to explore dormancy in three complementary ways. First, we will explore DCs formed by the genus Kurthia, which posses a unique cell envelope and morphogenesis program that provides new insights into bacterial evolution. Second, we will provide complete descriptions of the spore-like cells and mechanisms of dormancy in the three uncharacterized strains. Specifically, we will use electron cryotomography to describe the morphology of the specialized cell. We will also generate genomic, transcriptomic, and proteomic data, during entry and exit of the DC developmental program to get clues on the underlying genetic determinants that drive this process. Third using these genetic DC markers, we will identify new DC-forming species to demonstrate the widespread importance of this ecological survival strategy. Since most current detection methods used in molecular ecology are limited, we will develop a suite of novel methods to detect and study DC diversity in natural communities. We will extend the application of tailored metagenomic sequencing to identify the more abundant spore-forming species in the environment. The long-term goal is to develop a method for genomic sequencing of single spore-like cells in order to unveil rare DC- forming species in natural communities.
Our planet is expected to undergo increasing episodes of environmental stress. Given the primordial role of bacteria for ecosystem functioning, studying microbial dormancy as a model of
a natural response to changing environmental conditions could constitute a stepping-stone to improve our ability to predict the response of the biosphere to climate change.

