Bacterial rhythms and interactions
Rhythmic responses and competitive interactions in bacteria
Circadian timekeepers in environmental non-photosynthetic bacteria
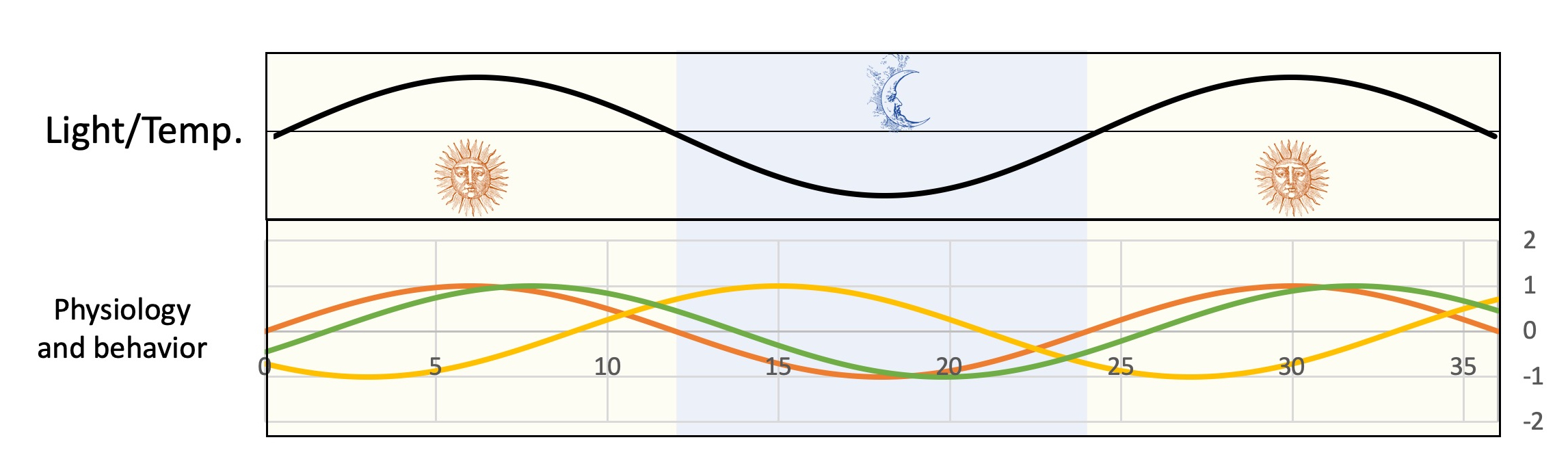
To coordinate their physiology with daily variations in light and temperature, Cyanobacteria have evolved a circadian clock, typically capable of entrainment (synchronization with the environment), persistance under constant conditions and temperature compensation (stability under a physiological temperature range). Preliminary evidence suggests that timekeeping mechanisms might exist in other, non-photosynthetic, bacteria and especially in some environmental, mostly plant-associated, Proteobacteria. This subproject aims at exploring the circadian behaviors of several bacteria and at identifying underlying molecular mechanisms. A further objective is to analyse clock-based rhythmic interactions within bacterial communities and between bacterial communities and multicellular hosts. Important practical consequences are expected if bacteria are found to use circadian clocks to synchronize with their host.
Rhythmic behaviors in aerobic anoxygenic phototrophic bacteria
Aerobic anoxygenic phototrophic bacteria (AANPs) are a phylogenetically diverse group of Gram negative bacteria which share the capacity to produce energy using cyclic photosynthesis. Although these bacteria are not strictly dependent on light for growth, their metabolism is strongly influenced by nycthemeral rhythms. How deeply other aspects of their lifestyle are affected by daily variations in light and temperature, however, is not known. This subproject aims at characterizing unsuspected rhythmic behaviors in AANPs including rhythmic interactions with other organisms and interferences between environmental and intrinsic cellular rhythms (cell cycle, DNA replication, cell division, etc.). In the process, we expect to demonstrate that coordination between different rhythms and, more generally, physiological timing is key to the life and success of a broad range of microorganisms.
Toxin-mediated competition in Escherichia coli
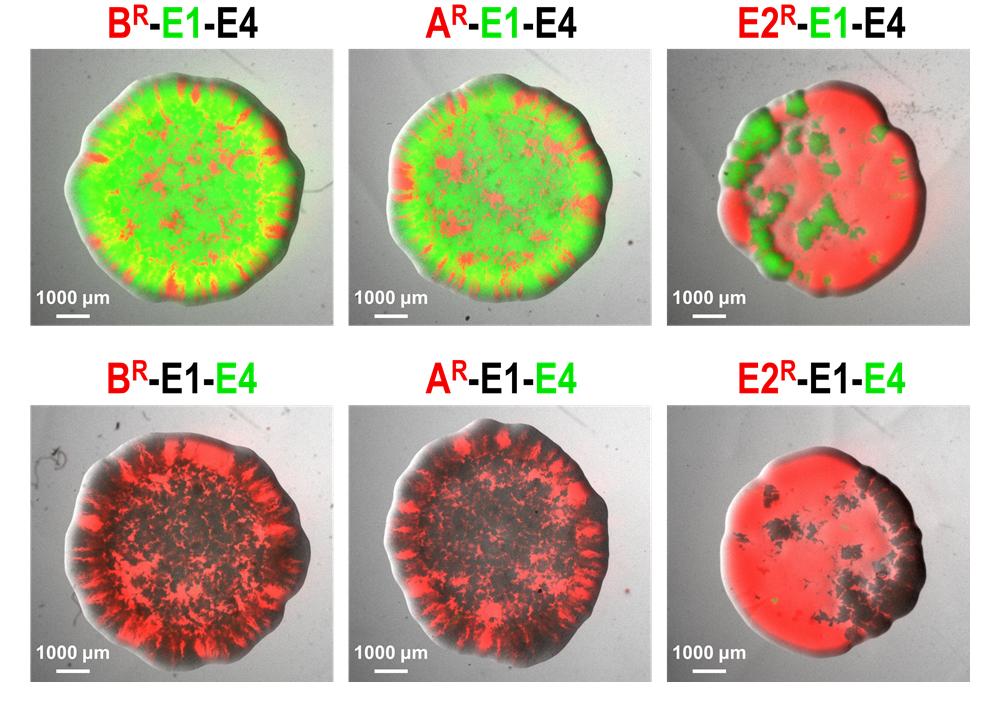
Microorganisms often live in dense communities where competition is fierce. To enhance their fitness, most bacteria produce toxins that inhibit or kill other bacteria. There is evidence that these antimicrobial compounds play a crucial role in the population dynamics of complex bacterial communities like the gut or the rhizosphere microbiota. Despite some precursory studies, detailed knowledge on how toxins may influence the succession of bacterial strains in such communities remains scarce. The main objective of this subproject is to characterize the strategies driving bacterial toxin production and connect them to particular ecologies. Through careful analysis of competitive interactions and modelling, we aim at better predicting bacterial population dynamics and evolutionary trajectories.
People involved in the project:
Diego Gonzalez (P.I.)
Bruno Cabete
Céline Terrettaz
Despoina Mavridou (Imperial College London, United Kingdom)
Martina Valentini (University of Geneva, Switzerland)

