Lab News
September 2020
Swiss National Science Foundation grant application funded. The SNF will provide 4 years of support for a research project linking lipogenesis and ascaroside signaling in the model organism C. elegans.
June 2020
Our manuscript describing the characterization of homo- and heterodimeric 2’- and 4’-isomeric ascaroside dimers from bacterivorous Caenorhabditis nematodes has been published in Organic Biomolecular Chemistry. We utilized a combination of HPLC-MS/MS precursor ion scanning, HR-MS/MS, and NMR techniques to identify a variety of dimeric ascarosides from Caenorhabditis remanei and Caenorhabditis nigoni. Structure assignments were confirmed by total synthesis of representative examples. Their biological functions were evaluated using a retention assay that indicated that males of C. remanei and C. nigoni are retained by their conspecific ascaroside dimers but not by the heterospecific isomers. These results demonstrate that dimerization of conserved monomeric ascaroside building blocks dramatically increases structural diversity and represents an efficient mechanism to generate species-specific ascaroside signals in the Caenorhabditis.
May 2020
A WormLab System from MBF Bioscience is installed at the Laboratory for Bioanalytical Chemistry to facilitate high-resolution worm imaging along with multiple worm tracking. The system will enable us to characterize the behavior modulating activities of novel secondary metabolites from nematode metabolomes.
March 2020
The lab goes into a state of hibernation for about six weeks due to COVID-19.
February 2020
Our highlight article focusing on the noncanonical biosynthesis of the homo-sesquiterpene sodorifen in the rhizobacterium Serratia plymuthica has been published as “Mechanism of the Month” in Trends in Chemistry.
February 2020
Marie-Desirée Scheidt joins the lab as a PhD student.
December 2019
The installation of a Bruker Ascend 600 NMR instrument at the Neuchatel Platform for Analytical Chemistry (NPAC) provides access to state-of-the-art high-resolution NMR spectroscopy for our research projects at the Laboratory for Bioanalytical Chemistry.
November 2019
Our manuscript describing the discovery of a novel glycolipid from the bacterivorous nematode Caenorhabditis nigoni has been published in Organic Letters. We utilized a combination of HPLC-MS/MS precursor ion scanning, HR-MS/MS, and NMR techniques to identify a side chain hydroxylated 4-epi-ascaroside that represents the first natural product carrying the L-3,6-dideoxy-lyxo-aldohexose unit, which we named caenorhabdose. Its structure was unambiguously established by total synthesis. The side-chain specificity of this 4-epi-ascaroside as well as those of 2-epi-ascarosides from Pristionchus pacificus suggests that nematodes generate species-specific glycolipids by epimerization of the conserved ascaroside core structures downstream from the ubiquitous peroxisomal β-oxidation cycle.
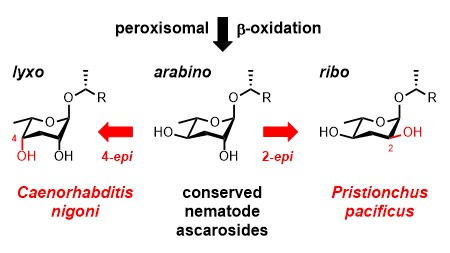
July 2019
Our manuscript on a novel class of modular fatty acid ascarosides from the bacterivorous nematode Caenorhabditis remanei has been published in Organic Letters. We utilized a combination of HPLC-MS/MS precursor ion scanning, micro-reactions, HR-MS/MS, MSn and NMR techniques to identify a series of fatty acid ascarosides that incorporate additional fatty acid building blocks. The structure of the dominating component called fasc#1 carrying a cyclopropyl fatty acid moiety was established by total synthesis. Biogenesis of this female produced male attractant depends on cyclopropyl fatty acid synthase (cfa), which is expressed in bacteria upon entering their stationary growth phase.
June 2019
Our manuscript describing the role of homologs of Caenorhabditis elegans chemosensory genes for behavior and chemotaxis in the root-knot nematode Meloidogyne incognita was published in Molecular Plant-Microbe Interactions. Using synthetic ascarosides from our lab the group of Uma Rao (ICAR-Indian Agricultural Research Institute, New Delhi, India) characterized behavioral responses in M. incognita and demonstrated their dependence on the chemosensory receptors odr-1, odr-3, tax-2, and tax-4.
March 2019
Our manuscript on guided cobamide biosynthesis for the heterologous production of reductive dehalogenases in organohalide-respiring bacteria has been published in Microbial Biotechnology. We utilized a combination of HPLC-HR-MS/MS and NMR techniques to identify novel cobamides that carry a purine or an 5-azabenzimidazol unit as lower base ligands. Their biogenesis was studied using media enriched with 15N ammonia or 15N labelled yeast extract in combination with MS techniques, which demonstrated that these heteroaromatic building blocks are derived from the yeast extract supplement. The research was performed in collaboration with Dr. Torsten Schubert and Prof. Gabriele Diekert at the Department of Applied and Ecological Microbiology, Institute of Microbiology, Friedrich Schiller University, in Jena, Germany.
August 2018
Our manuscript on the biogenesis of polymethylated sodorifen in the rhizobacterium Serratia plymuthica has been published in the Journal of the American Chemical Society (JACS). We utilized a combination of comparative GC-MS and NMR analysis of wild-type and mutant strains along with in vitro assays with heterologously expressed enzymes to demonstrate that sodorifen biogenesis involves methylation and cyclization of the canonical farnesyl pyrophosphate by a C-methyltransferase (MT) to provide monocyclic pre-sodorifen pyrophosphate as a substrate for the terpene cyclase (TC). Furthermore, we employ in vivo feeding experiments with 13C labelled precursors along with NMR spectroscopy to elucidate the biogenetic pathway from farnesyl pyrophosphate via pre-sodorifen pyrophosphate to sodorifen. The research was performed in collaboration with Prof. Birgit Piechulla and her team at the University of Rostock, Germany.
May 2018
A review article describing our research focusing on ascaroside glycolipids as key signaling molecules in nematodes has been published in CHIMIA.
May 2018
Dr. Bandi Siva joins the lab as a Postdoctoral Researcher.
March 2018
Our manuscript describing comparative ascaroside profiling of various Caenorhabditis species and the characterization of species-specific (ω) and (ω – 2)-hydroxylation of ascaroside aglycones downstream of the peroxisomal β-oxidation cycle has been published in a special issue of the Journal of Organic Chemistry (JOC) dedicated to the synthesis of antibiotics and related molecules. The research was performed in collaboration with Prof. Jagan Srinivasan and Douglas K. Reilly from Worcester Polytechnic Institute (WPI), Massachusetts, USA.
September 2017
Our manuscript describing a novel GC-EIMS-based ascaroside screen for crude nematode exo-metabolomes is accepted for publication in Analytical Chemistry (ACS).
May 2017
Franziska Dolke receives her PhD degree (Dr. rer. nat.) from the Friedrich Schiller University in Jena, Germany, for her research on ascaroside components in Caenorhabditis species.
March 2017
Chuanfu Dong receives his PhD degree (Dr. rer. nat.) from the Friedrich Schiller University in Jena, Germany, for his research on ascaroside components in Caenorhabditis species.
Rocío Rivera Sanchez joins the lab as a PhD student.
First worms grown in the nematode lab.
February 2017
Célia Bergame joins the lab as a PhD student.
September 2016
Swiss National Science Foundation grant application funded. The SNSF will provide support for a research project focusing on the comparative analysis of secondary metabolism in bacterivorous nematodes.
August 2016
Stephan is honored to deliver the plenary lecture during the 32nd symposium of the European Society of Nematologists (ESN) in Braga, Portugal
The Laboratory of Bioanalytical Chemistry is installed at the University of Neuchâtel.