Dr. Thomas Badet

Maître Assistant (Lecturer)
Bureau A313
University of Neuchâtel
Rue Emile-Argand 11 | CH-2000 Neuchâtel | Switzerland
+41 (0) 32 718 22 05 | thomas.badet@unine.ch
Teaching activities
(i) Cours d’« Introduction à la phytopathologie » (3BT1010) du semestre de printemps pour le BSc de biologie – 28h
Méthode : cours ex cathedra, discussions.
Support : diapositives comme support visuel, Moodle.
Évaluation : contrôle écrit en fin de semestre pour un total de 3 ECTS.
(ii) Cours de « Plant Pathology » (3BL2206) du semestre de printemps pour le MSc de biologie – 28h
Méthode : mélange de cours ex cathedra, exercices, exposés par les étudiants.
Support : diapositives comme support visuel, Moodle.
Évaluation : contrôle continu basé sur la participation, une présentation d’article individuelle et un projet de recherche présenté en groupe pour un total de 3 ECTS.
(iii) Cours de “Bioinformatics tools” (3BL2194) du semestre d’automne pour MSc de biologie – 30h
Méthode : mélange de cours ex cathedra et exercices.
Support : diapositives comme support visuel, Moodle.
Évaluation : contrôle continu basé sur des exercices notés pour un total de 3 ECTS.
(iiii) Apprentissage par problème (APP 3BL1171) en biologie moléculaire et cellulaire pour le BSc de biologie – cours bloc
Méthode : cours bloc, discussions.
Support : Moodle.
Évaluation : rapport écrit et interview pour un total de 9 ECTS.
Research focus
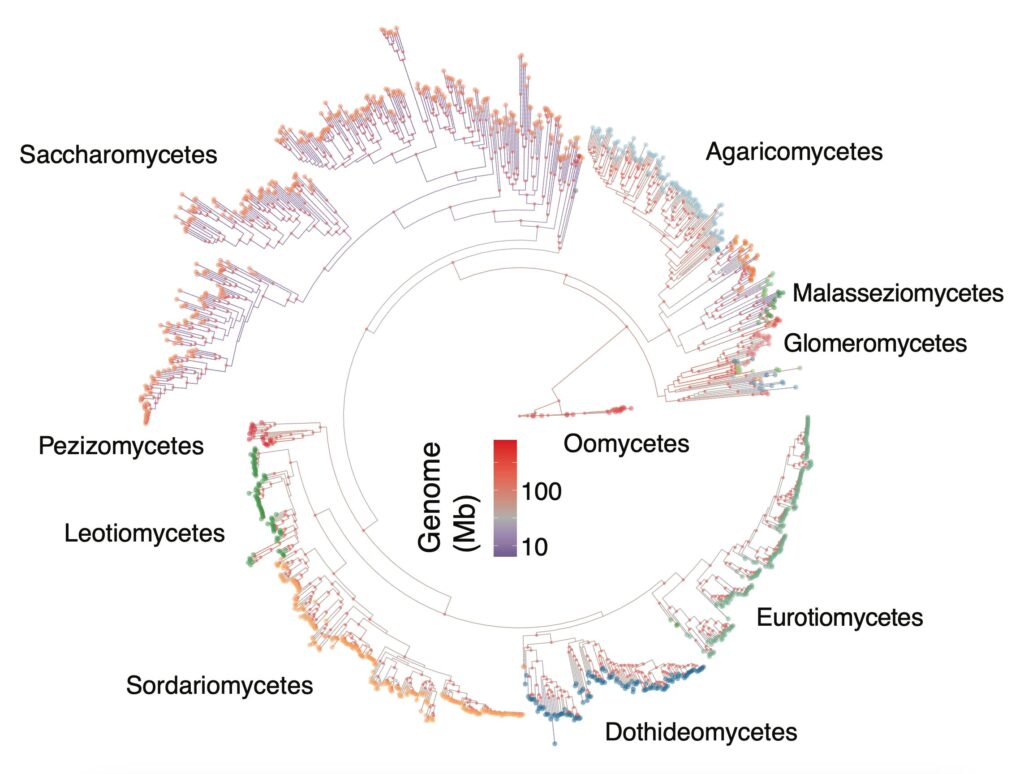
Fungal pathogens’ pangenomics. Modern agricultural practices rely on the extensive use of fungicides for disease control. If fungicides are an effective measure against pathogens when first introduced, history has told us that their efficiency rapidly decreases as resistant individuals emerge in field populations (Fisher et al., 2018; Lucas et al., 2015). Genome diversity is of particular importance in the context of rapid adaptation. Moving away from the “single reference genome” paradigm is a prerequisit for better understading what drives pathogen’s evolution in the context of crop production. Indeed, the gene content of a species largely governs its ecological interactions and adaptive potential. A species is therefore defined by both core genes shared between all individuals and accessory genes segregating presence-absence variation. Part of my research focus on accessing the extent of fungal pathogens intraspecific genomic diversity through the construction of pangenomes.
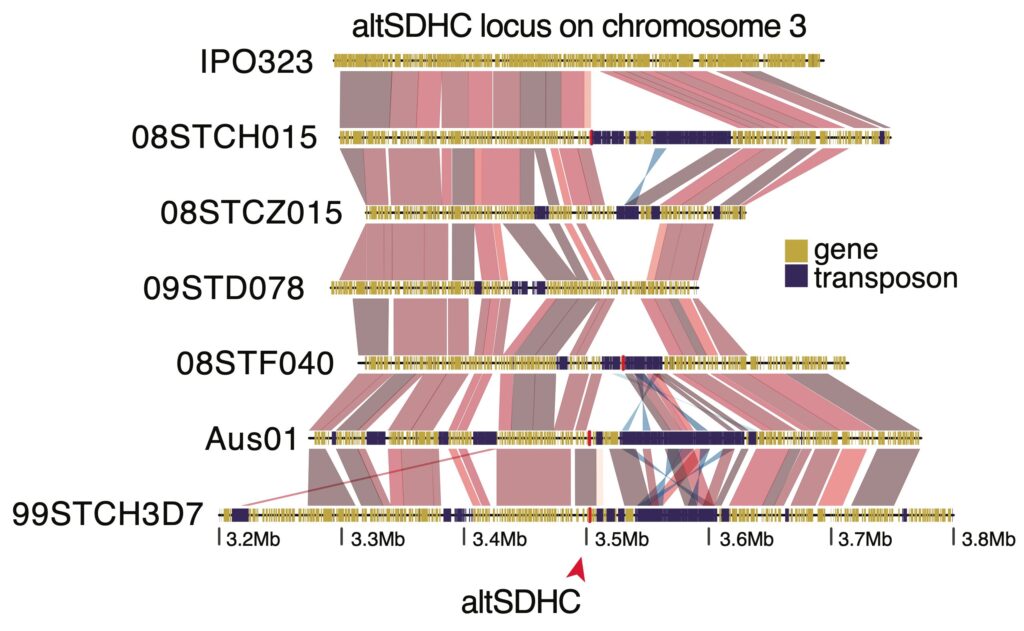
Transposable elements and genome defenses in fungi. Fungi harbor some of the highest mutation rate outside from viruses. Fungal exceptional mutation rate is due to a defense mechanism against genomic parasites known as transposable elements. The process is known as Repeat-Induced Point mutations (RIP) and likely holds a central role in genome evolution. RIP takes place upon sexual recombination, after fertilization and before karyogamy, inducing multiple C-to-T mutations across both duplicated sequences (refs). The wave of RIP mutations occurring upon a single sexual cycle leads to the disruption of the coding sequence, turning transposons inactive. Despite the prevalence of RIP, transposable elements often constitute a large fraction of fungal genome. I’m currently investigating the distribution of signatures of RIP mutations and their impact on genome architecture across the fungal kingdom. Focusing on Neurospora crassa, I also aim at characterizing the genes controlling the mechanism of RIP mutation.
Short CV
2021-Today > Lecturer part of the laboratory of Prof. Vermeer (University of Neuchâtel, Switzerland)
2018-Today > Scientific Collaborator part of the laboratory of Prof. Croll (University of Neuchâtel, Switzerland)
2014-2017 > PhD in Plant Pathology with Dr. Raffaele (INRA Toulouse, France)
2012-2014 > MSc in Plant Biology (University of Toulouse, France)
Publications (Google scholar link)
2024
A systematic screen for co-option of transposable elements across the fungal kingdom
U Oggenfuss, T Badet, D Croll
Mobile DNA 15, 2 (2024)
2023
Recent reactivation of a pathogenicity-associated transposable element is associated with major chromosomal rearrangements in a fungal wheat pathogen
T Badet, SM Tralamazza, A Feurtey, D Croll
Nucleic Acids Research, (2023)
2021
Rapid sequence evolution driven by transposable elements at a virulence locus in a fungal wheat pathogen
NK Singh, T Badet, L Abraham, D Croll
BMC genomics 22 (1), 1-16 (2021)
A population-level invasion by transposable elements triggers genome expansion in a fungal pathogen
U Oggenfuss, T Badet, T Wicker, FE Hartmann, NK Singh, L Abraham, et al,
Elife 10, e69249 (2021)
Machine-learning predicts genomic determinants of meiosis-driven structural variation in a eukaryotic pathogen
T Badet, S Fouché, FE Hartmann, M Zala, D Croll
Nature communications 12 (1), 1-14 (2021)
Emergence and diversification of a highly invasive chestnut pathogen lineage across southeastern Europe
L Stauber, T Badet, A Feurtey, S Prospero, D Croll
Elife 10 (2021)
2020
A 19-isolate reference-quality global pangenome for the fungal wheat pathogen Zymoseptoria tritici
T Badet, U Oggenfuss, L Abraham, BA McDonald, D Croll
BMC biology 18 (1), 1-18 (2020)
The rise and fall of genes: origins and functions of plant pathogen pangenomes
T Badet, D Croll
Current opinion in plant biology 56, 65-73 (2020)
Stress-driven transposable element de-repression dynamics and virulence evolution in a fungal pathogen
S Fouché, T Badet, U Oggenfuss, C Plissonneau, CS Francisco, D Croll
Molecular Biology and Evolution 37 (1), 221-239 (2020)
2019
Expression polymorphism at the ARPC4 locus links the actin cytoskeleton with quantitative disease resistance to Sclerotinia sclerotiorum in Arabidopsis thaliana
T Badet, O Léger, M Barascud, D Voisin, P Sadon, R Vincent, A Le Ru, et al,
New Phytologist 222 (1), 480-496 (2019)
2017
Parallel evolution of the POQR prolyl oligo peptidase gene conferring plant quantitative disease resistance
T Badet, D Voisin, M Mbengue, M Barascud, J Sucher, P Sadon, et al,
PLoS genetics 13 (12), e1007143 (2017)
Codon optimization underpins generalist parasitism in fungi
T Badet, R Peyraud, M Mbengue, O Navaud, M Derbyshire, RP Oliver, et al,
Elife 6, e22472 (2017)
2016
Emerging Trends in Molecular Interactions between Plants and the Broad Host Range Fungal Pathogens Botrytis cinerea and Sclerotinia sclerotiorum
M Mbengue, O Navaud, R Peyraud, M Barascud, T Badet, R Vincent, et al,
Frontiers in plant science 7, 422 (2016)
2015
Common protein sequence signatures associate with Sclerotinia borealis lifestyle and secretion in fungal pathogens of the Sclerotiniaceae
T Badet, R Peyraud, S Raffaele
Frontiers in plant science 6, 776 (2015)
2014
Resistance to phytopathogens e tutti quanti: placing plant quantitative disease resistance on the map
F Roux, D Voisin, T Badet, C Balagué, X Barlet, C Huard‐Chauveau, et al,
Molecular plant pathology 15 (5), 427 (2014)